A new open-source coronavirus simulation, released to help authorities forecast the effects of local lockdown measures, has predicted that a second wave of COVID 19 is probable in ‘almost all cases’ across the London Boroughs for which it has been run, although the outbreak is likely to be significantly less severe than the first.
The freely available tool, which has already been used for decision making by authorities in North London, forecasts how localised lockdown measures – such as closing a local shop, or quarantining the residents of a particular building – affect the spread of coronavirus.
The modellers behind it, who have described their simulation as ‘pessimistic,’ found a second wave of coronavirus is likely, regardless of the measures put in place.
“The idea that your local Morrisons will be featured in a simulation is very uncommon,” said Dr Derek Groen, a lecturer in simulation and modelling at Brunel University London.
“Our simulations allow authorities to see the impact of things like closing a local supermarket, or changing the constraints placed on schools or businesses at a very local level.”
The simulation was validated up to 80 days by Brunel’s Dr David Bell using anonymised data supplied by the London North West University Healthcare NHS Trust and contains a “very wide range of local intervention types” – from shutting pubs or limiting access to parks, to closing certain businesses or restricting individual households – to help local authorities plan lock-down measures.
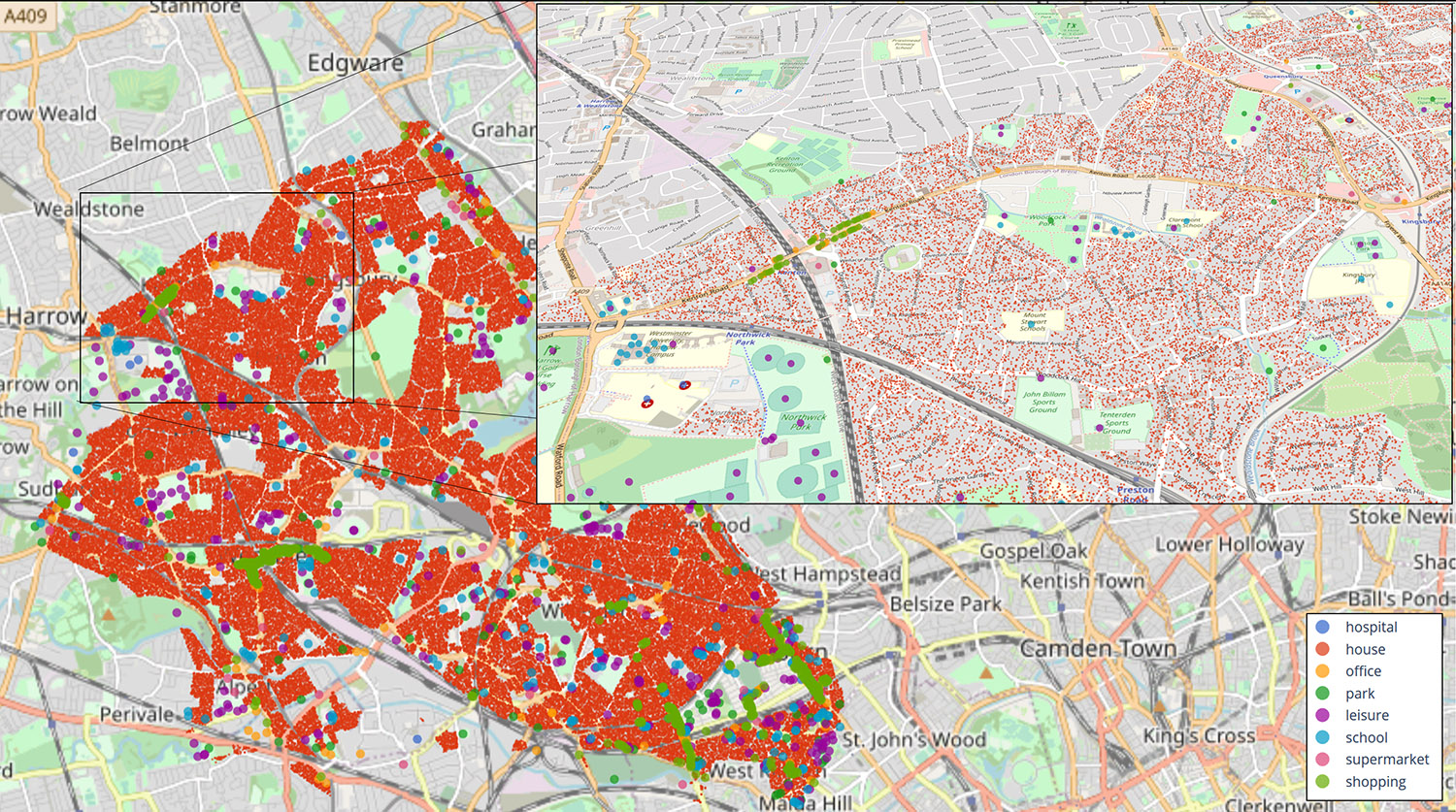
Example location map
Full simulations have already been developed for a number of London boroughs, including Brent, Ealing, Hillingdon, and Harrow, and more basic forecasts have been produced for Westminster, Kensington, Fulham and Chelsea.
The full simulations considered several scenarios – including that an area takes no measures, that an area enters an extended lockdown period or that an area runs a ‘dynamic’ lockdown using a variety of changing interventions – with all of the lockdown scenarios showing a marked decrease in local infections.
The simulation allows infection rates to be visualised on a map, run using OpenStreetMaps, letting modellers see where hotspots develop, and how hotspots move as the disease spreads through a population.
“The results between boroughs has so far varied a lot, so we’re trying to understand better why that is,” said Dr Groen.
“The models definitely see a second wave in almost all cases, although it looks much less deep than the first wave, but it could be more prolonged.”
The simulations are based on assumptions drawn from scientific literature, such as how infectious each patient is and how many people adhere to social distancing rules, although the open source nature of the code means that anyone using it can update those assumptions as further research becomes available, the researchers say.
“So, for example, it’s not totally clear at the moment how many people are wearing masks,” said Dr Groen. “We can put in a number, say, 20%, then run the simulation – but if people want to put in a different number, they can just do more runs and vary the number.”
The simulations have already been used by one local authority in North London, the researchers say, and its now hoped that further local authorities will download the tool to support their own decision making.
Dr Imran Mahmood, a Research Fellow at Brunel, who co-created the simulations said: “The local authority had been considering what changes they needed to make regarding bed capacity in the coming years, and whether they should redirect patients to other hospitals, and these models helped support those decisions.
“We hope that others can take the codes and model their local areas – so councils could do that or even local volunteers, to get an idea of how COVID-19 is spreading in their local community.
“It probably can’t be set up by a complete layman, but people familiar with building simulations could do it.”
The Flu And Coronavirus Simulator (FACS) was made possible through a collaborative effort by the Department of Computer Science at Brunel and the HiDALGO project (http://hidalgo-project.eu), and is available to download now from GitHub: https://github.com/djgroen/facs
The FACS team are: Dr Imran Mahmood, Dr Hamid Arabnejad, Dr Diana Suleimenova, Dr Isabel Sassoon, Dr Alaa Marshan, Dr Alan Serrano, Prof Panos Louvieris, Dr Anastasia Anagnostou, Prof Simon J E Taylor, Dr David Bell, Dr Derek Groen
For further information on Computer Science at Brunel, please visit: brunel.ac.uk/computer-science
Reported by:
Tim Pilgrim,
Media Relations
+44 (0)1895 268965
tim.pilgrim@brunel.ac.uk