Serial Re-sequencing of Evolving Populations
The use of ‘selection-resequencing’ (SR) experiments has been increasing with the application of next-generation sequencing methodologies. We have developed our own experimental approach and analysis resources in order to address specific issues:
- The avoidance of bottlenecks to address adaptation at a population level and to maximize sensitivity to identify useful changes
- The inclusion of replicate independent cultures
- The inclusion of controls
- The ability to distinguish between process-adaptations to the experiment and the intended target behaviour adaptations
- The detection of mutations that confer fitness advantages but which due to clonal interference and serial clonal competition do not emerge to dominance
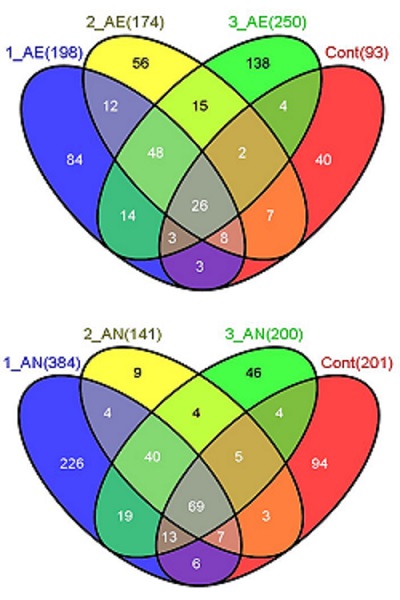
The findings of studies using this methodology are being used in commercially sensitive projects that are not currently published. However, this methodology is working well, identifying both specific and general areas for strain improvement for fermentation applications. Illustrative outcomes in terms of selected mutations are shown in the figure below, which shows the number of mutations seen in a total of 96 resequencing datasets, addressing three independent lineages and controls, for aerobic and anaerobic culture conditions associated with tolerance to 2,3-butanediol.
Points to note:
- 26 and 69 of the mutations (grey) occur in all lineages, including controls – emphasizing the need for controls to identify non-specific experimental selection
- 48 and 40 of the mutations (mid green) occur in all three independent lineages, and not in the controls